Protein Structure & Bioinformatics: MCQ and Short Answer Review (Academic & Detailed)
Question 1
Generally, the function of a protein depends on its: (a) One-dimensional structure (b) Two-dimensional structure (c) Three-dimensional structure (d) Four-dimensional structure
Correct Answer: (c) Three-dimensional structure Explanation: A protein's specific biological activity and function (e.g., binding a substrate, acting as an enzyme, or transporting a molecule) is uniquely determined by its final, folded shape, which is its tertiary structure (3D structure).
Question 2
The protein domains are: (a) Functional and structural units within protein (b) Secondary structural elements (c) Linear sequences of amino acids (d) Specific regions for post-translational modification
Correct Answer: (a) Functional and structural units within protein Explanation: Protein domains are distinct, compact, and independently folding regions of a polypeptide chain. They are considered both structural units and often functional units, with each domain performing a specific task.
Question 3
The first step in x-ray crystallography experiment is: (a) Compute an electron density (b) Build a model of your molecule (c) Measure a diffraction pattern (d) Grow a crystal
Correct Answer: (d) Grow a crystal Explanation: X-ray crystallography requires the protein molecules to be highly ordered in a crystal lattice so that they diffract the X-rays coherently. Therefore, growing a pure crystal of the protein is the essential first step before diffraction patterns can be measured.
Question 4
What is primary role of computational biology? (a) Using computer algorithms to analyze data (b) Identifying genetic mutations (c) Studying protein functions (d) Analyzing the expression patterns
Correct Answer: (a) Using computer algorithms to analyze data Explanation: Computational biology is an interdisciplinary field that focuses on developing and applying computer algorithms and mathematical models to analyze and interpret large, complex biological data sets (genomic, proteomic, etc.). Options (b), (c), and (d) are examples of what computational biology is used for, not its primary role.
Question 5
Which computational approach is used to predict protein structure based on amino acid sequence? (a) Multiple sequence alignment (b) Homology modelling (c) Clustering analysis (d) BLAST searches
Correct Answer: (b) Homology modelling Explanation: Homology modelling (or comparative modeling) is the method that constructs a 3D model for an unknown protein based on the experimentally determined structure of a related (homologous) protein template. This method requires the input of the amino acid sequence.
SECTION 2: SHORT QUESTIONS (Short Answers and Definitions)
1. Define domains of the protein.
A protein domain is a fundamental, compact, three-dimensional structural and functional unit within a protein. Each domain folds independently into a stable structure and often carries out a specific function, such as binding to DNA, binding to a small molecule, or possessing enzymatic activity. Large proteins are frequently composed of multiple distinct domains, suggesting they evolved through the shuffling of these modular units.
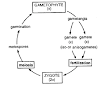
2. How corona virus enters the host cells?
The Coronavirus (specifically SARS-CoV-2) enters host cells through a series of steps:
Attachment (Binding): The viral Spike (S) protein, located on the surface of the virus, binds to the ACE2 receptor (Angiotensin-Converting Enzyme 2) found on the membrane of host cells (e.g., in the lungs and heart).
Activation (Cleavage): Host cell proteases, such as TMPRSS2, cleave the Spike protein, a step necessary to activate it for the fusion process.
Membrane Fusion: The activated Spike protein mediates the fusion of the viral envelope with the host cell membrane.
Entry: This fusion releases the viral genetic material (RNA) into the host cell's cytoplasm, initiating the infection cycle.
3. Define genomics.
Genomics is the comprehensive study of the structure, function, evolution, mapping, and editing of entire genomes. A genome includes all of an organism's genetic material. Genomics utilizes high-throughput DNA sequencing, bioinformatics, and computational methods to understand the complete genetic makeup and its influence on the organism's traits and health.
4. Differentiate between genomics and proteomics.
The key difference lies in the molecules they study:
5. What is GenBank. Describe it briefly.
GenBank is the NIH (National Institutes of Health) genetic sequence database—a publicly accessible, comprehensive repository that contains all publicly available sequences and their associated descriptive information (annotations).
Brief Description:
It is maintained by the NCBI (National Center for Biotechnology Information).
Data is submitted by scientists and sequencing centers across the globe.
It is part of a global consortium, ensuring daily exchange of sequence data with other major international databases.
GenBank is a cornerstone of bioinformatics, enabling researchers to perform sequence alignments, identify gene homologs, and study evolutionary relationships.
6. Write a short note on Protein Data Bank (PDB).
The Protein Data Bank () is the central, worldwide archive for information concerning the three-dimensional (3D) structures of large biological macromolecules (proteins, DNA, and RNA).
Short Note:
Source Data: PDB structures are primarily determined through experimental methods like X-ray Crystallography, Spectroscopy, and Cryo-Electron Microscopy (Cryo-).
Content: It stores the 3D atomic coordinates, making the structures available for visualization and analysis.
Application: The PDB is vital for structural biology and drug design. By providing the detailed shape of biological targets, it allows researchers to understand molecular mechanisms and design new drugs that precisely fit into and modify the activity of target proteins (like enzyme inhibitors).




+by+Blacklotus+Landscaping.jpg)


.jpg)


0 Comments